Slicer学习笔记(十六)分割配准
-
-
- 1、记录配准过程:
- 2、尝试不同的配准方法,找最好配准效果的方法
-
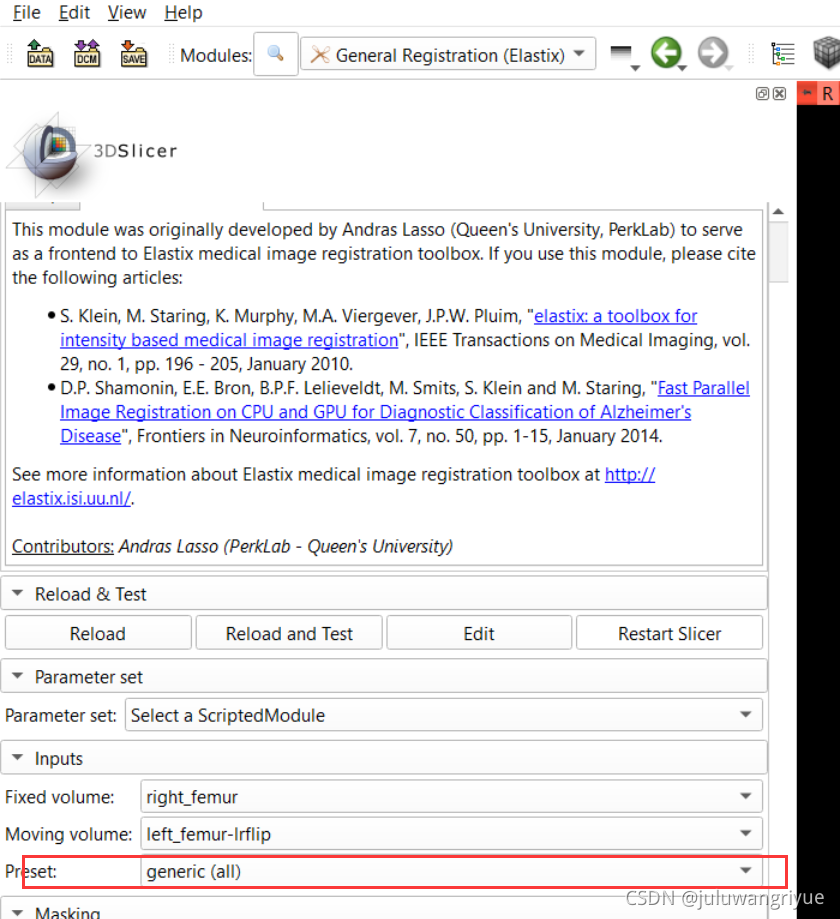
- generic rigid (all)
- 3D MR T1, monomodal (brain)
- 3D MR BFFE, monomodal (prostate)
- 3D MRT1 & PET (brain)
- 3D CT, monomodal (lung)
- 3D MRI, monomodal (brain)
- 3D MR HASTE, monomodal (brain)
- 2D x-ray- 3D CT (cerebral)
- 3D microCT (whole-body mouse)
- 3D MR, monomodal (knee cartilage)
- 3D MR, multi sequence (carotid artery)
- 3D CT, monomodal (head and neck)
- 3D MR, multi-contrast (rat brain)
- 3D CT, 3D MR, multimodal (head and neck)
- 3D MR, multi-contrast (mouse brain)
- 3D T1 fat-suppressed Ga-enhanced extremity MR (wrist)
- 3D DCE-MRI (breast)
- 3D MR (mouse brain)
-
- 3、slicer中Elastix配准的实现
1、记录配准过程:
1、将一个CT切分成左侧和右侧两个部分,因为关心的是髋骨部分,所以就取了CT中的一部分用于配准。
2、因为此次任务是用健康的股骨替换磨损的股骨部分,此CT中左侧股骨是健康的,右侧股骨是磨损的,所以用右侧股骨为固定图像(Fixed volume),用左侧股骨为浮动图像,最终生成配准图像用于替换右侧磨损的图像。
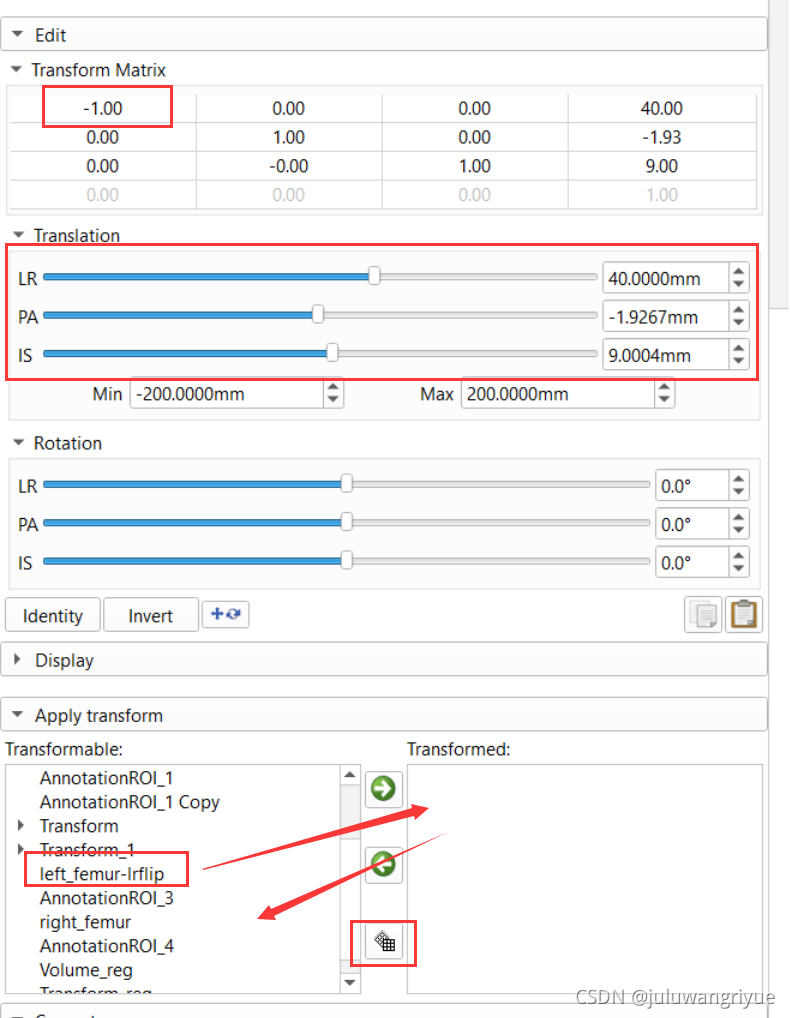
3、对于切分的左侧图像,第一步是变换成与右侧图像相同的状态,需要做的是 x轴镜像,需要在slicer中选择 Transforms模块,通过修改Transform Matrix 的x轴参数1改成-1 实现x轴镜像,因为是 − 1 ∗ p x -1*p_x −1∗px 即所有x都变成了相反数,原体素空间位置 ( x , y , z ) (x,y,z) (x,y,z) 变成了 ( − x , y , z ) (-x,y,z) (−x,y,z) 整体看起来数据关于原点对x轴做了镜像。然后是x、y、z轴的平移,使镜像后的数据与右侧股骨数据基本对齐,可能不对齐也可以使用。
4、在下图Transformable中选择需要变换的数据,使用向右的箭头添加到Transformed中,确定变换后,点击向左箭头下方的 Harden transform图标。此时图像会生效变换,并自动跳回左侧。
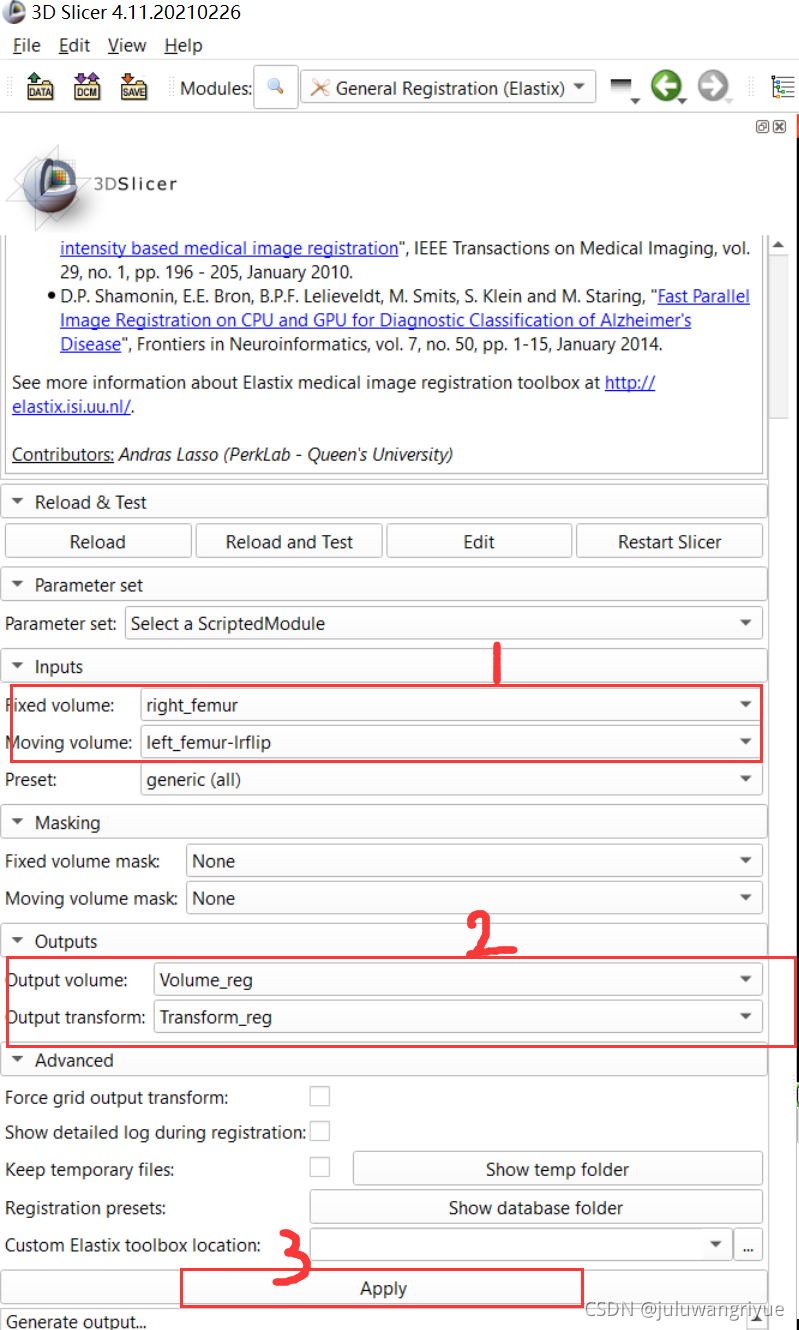
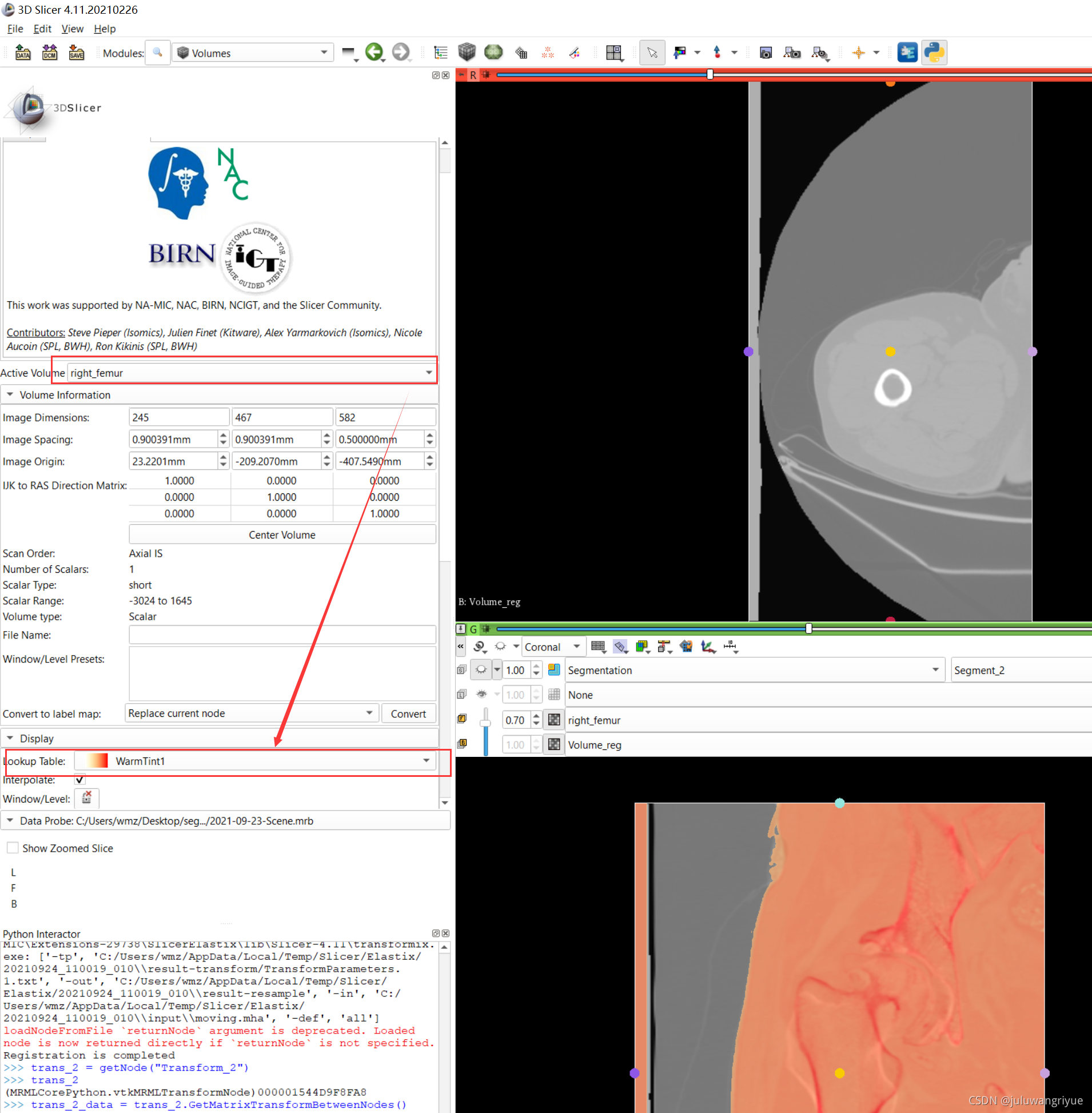
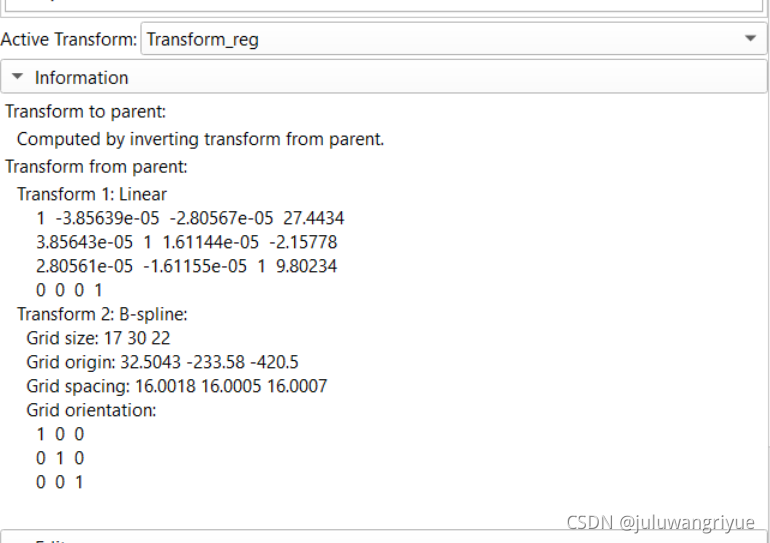
5、此时再使用 General Registration(Elastix) 模块实现配准变换。选定 Fixed volume: right_femur,Moving volume: left_femur-lrflip和 输出自己命名两个变量Volume_reg和Transform_reg,然后执行 Apply,等待执行结束就可以了。
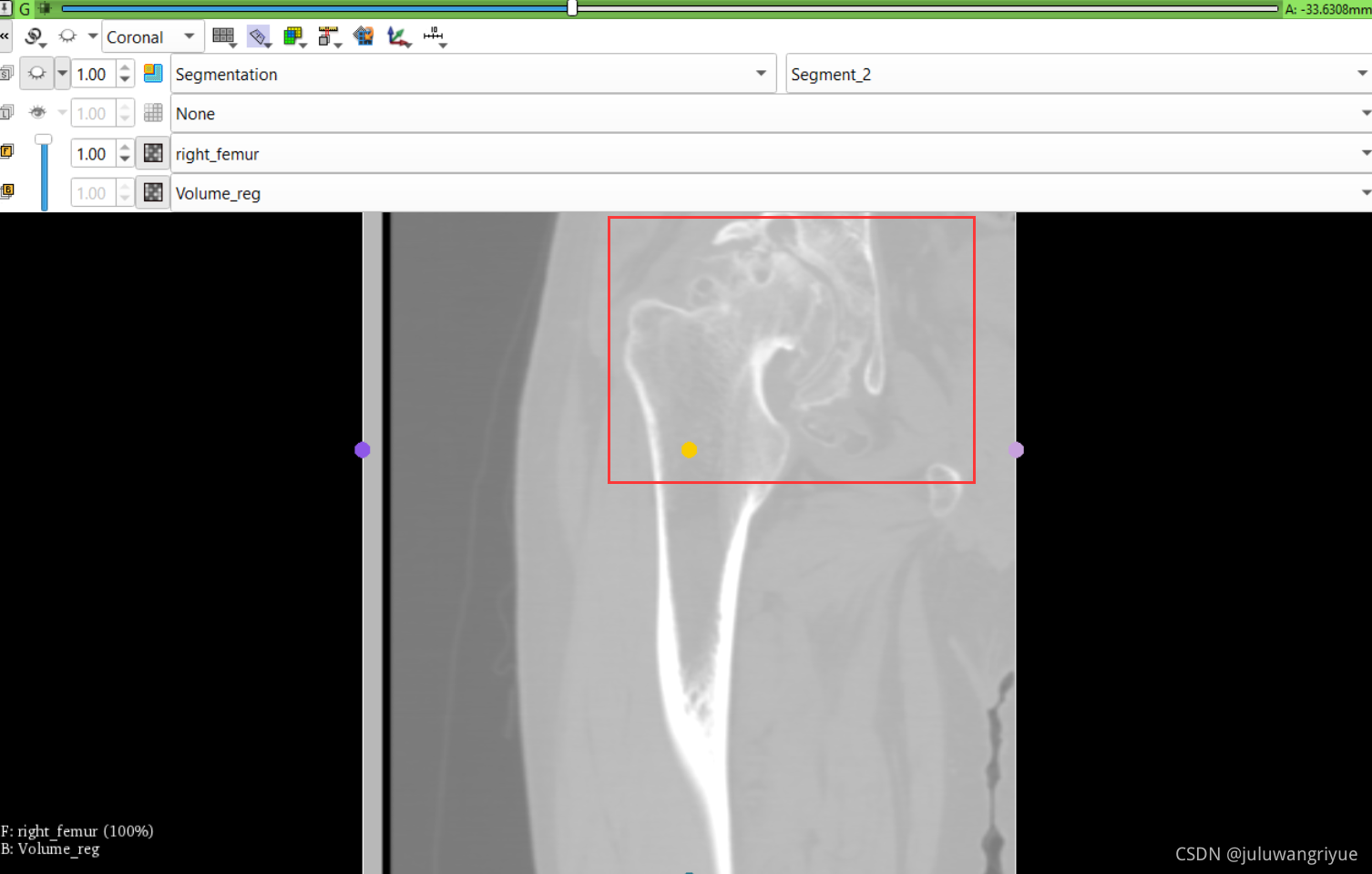
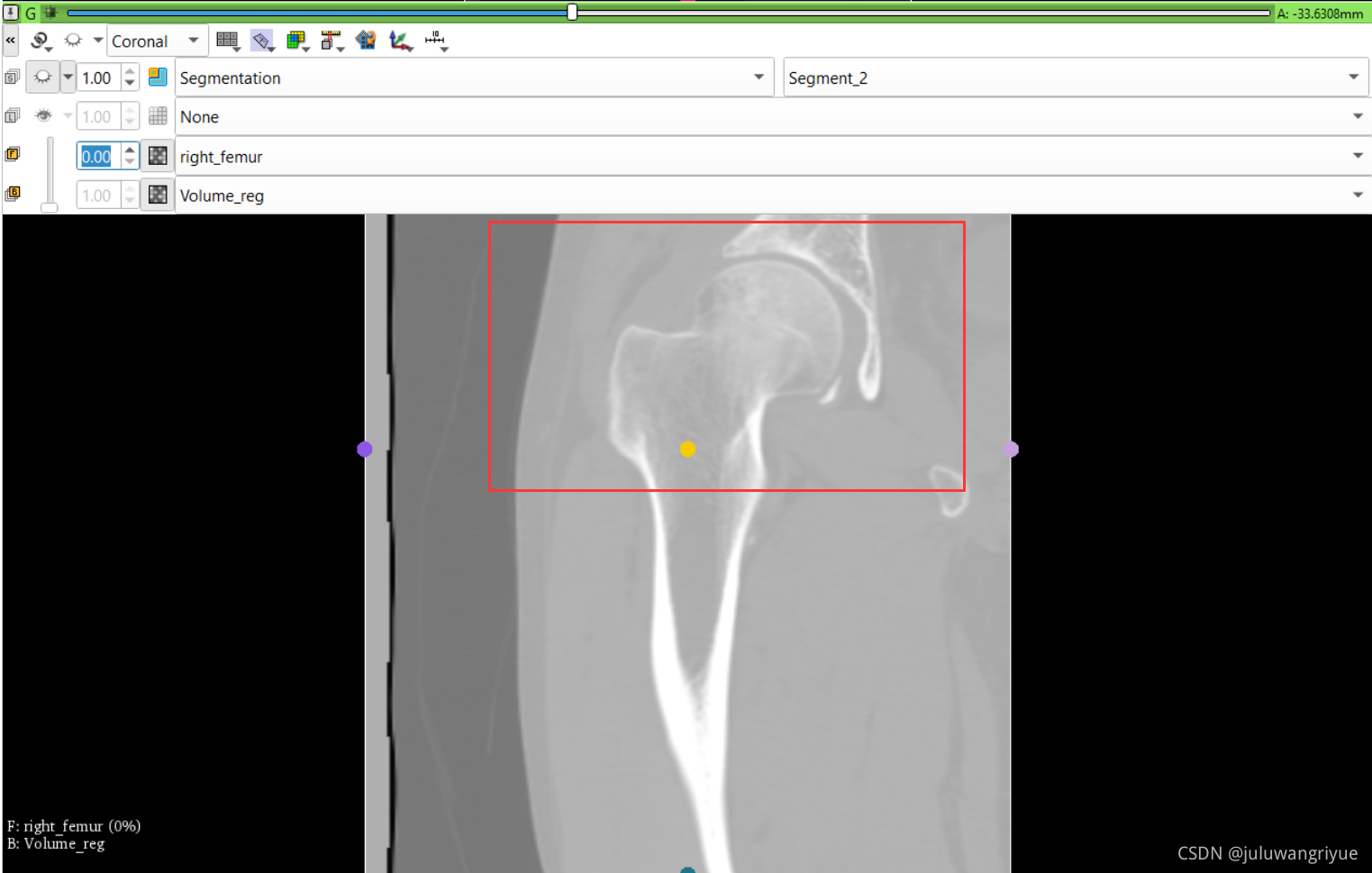
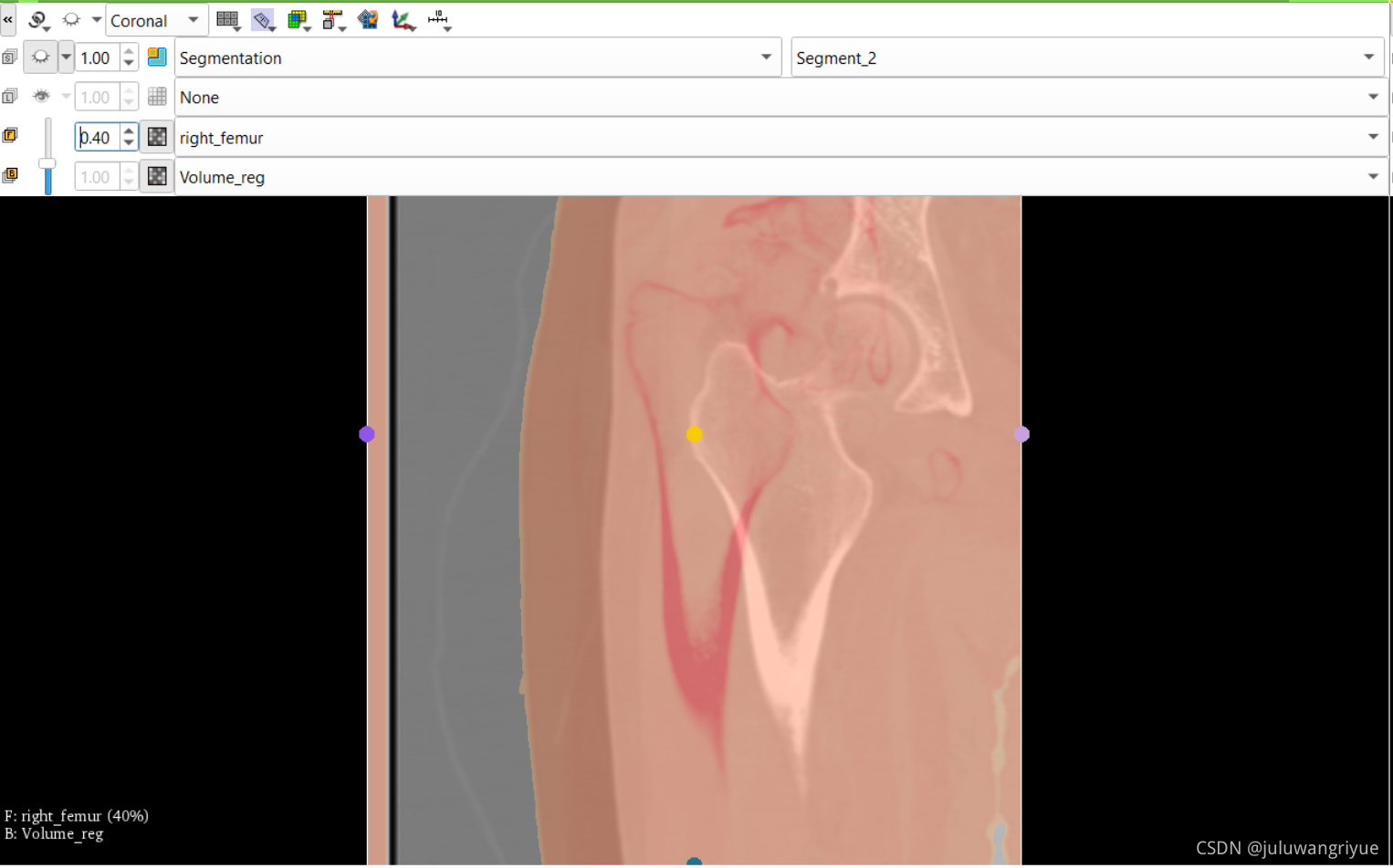
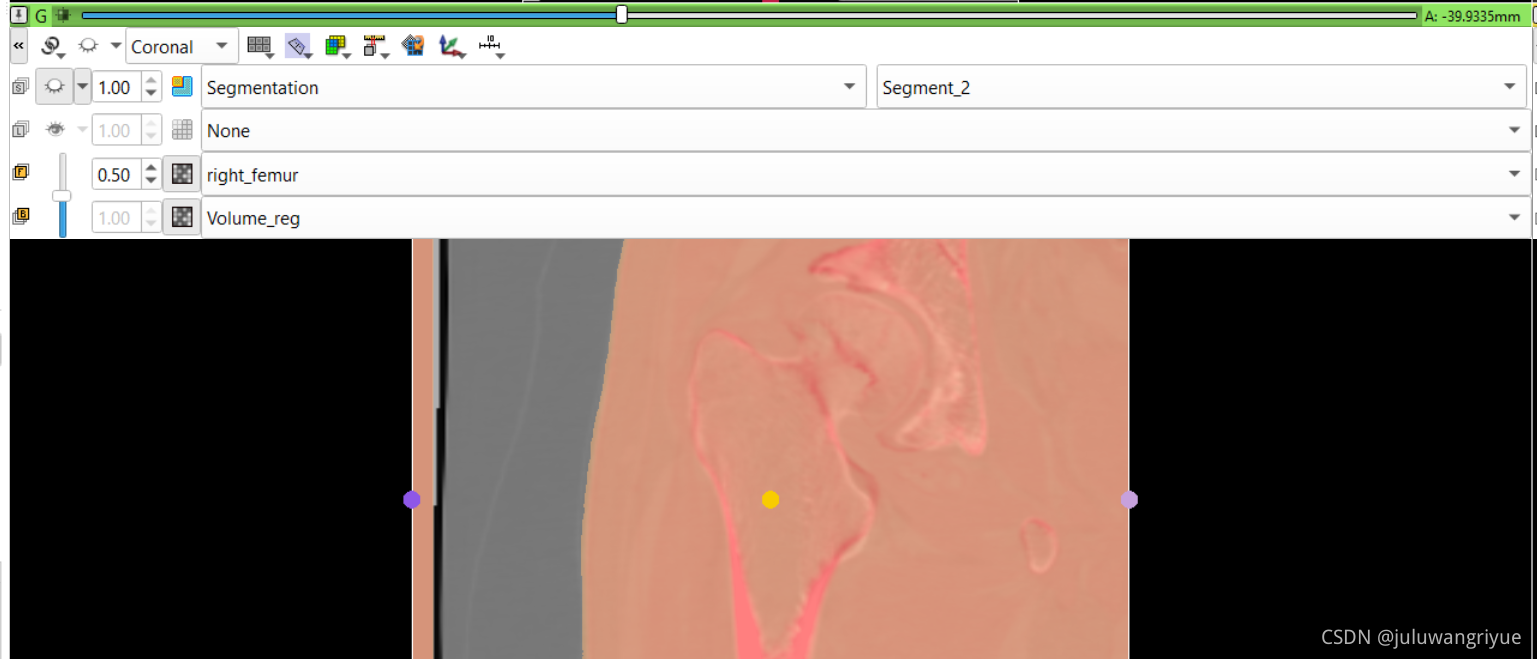
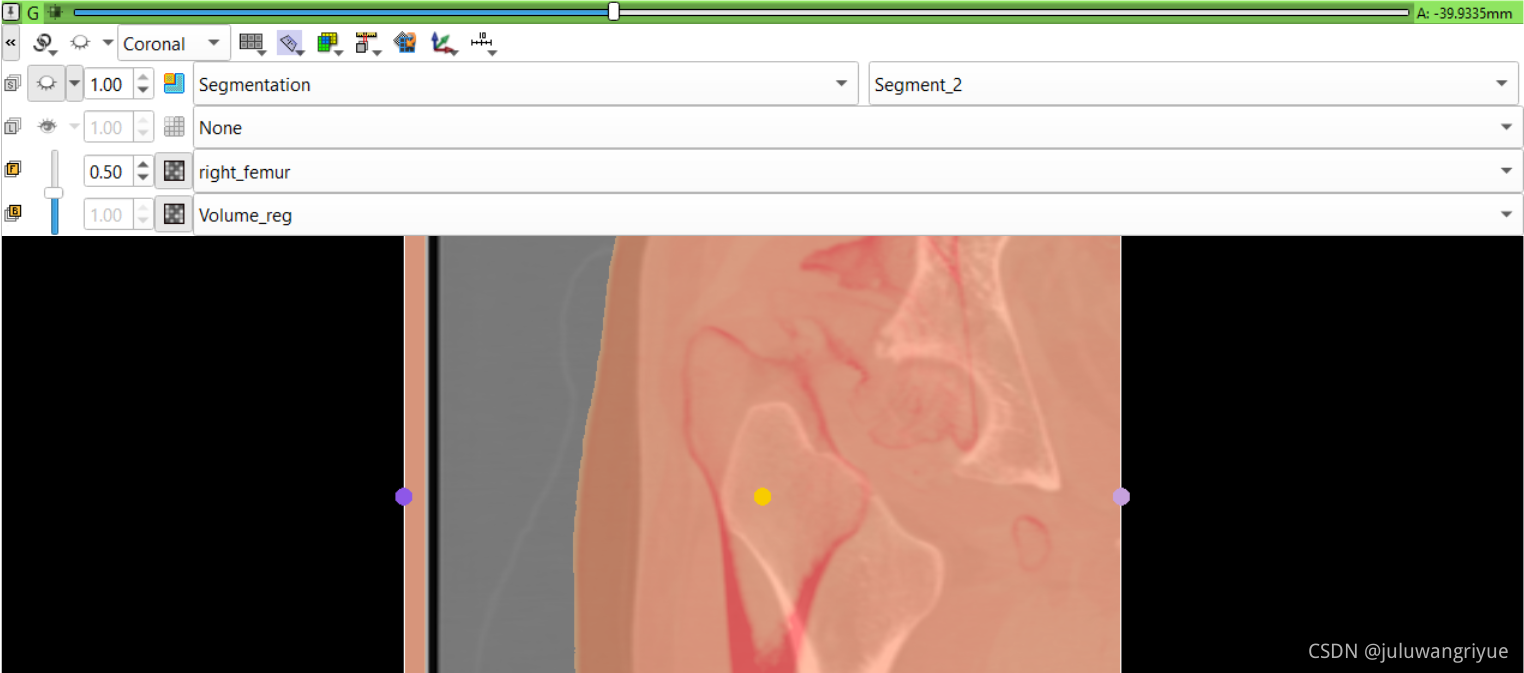
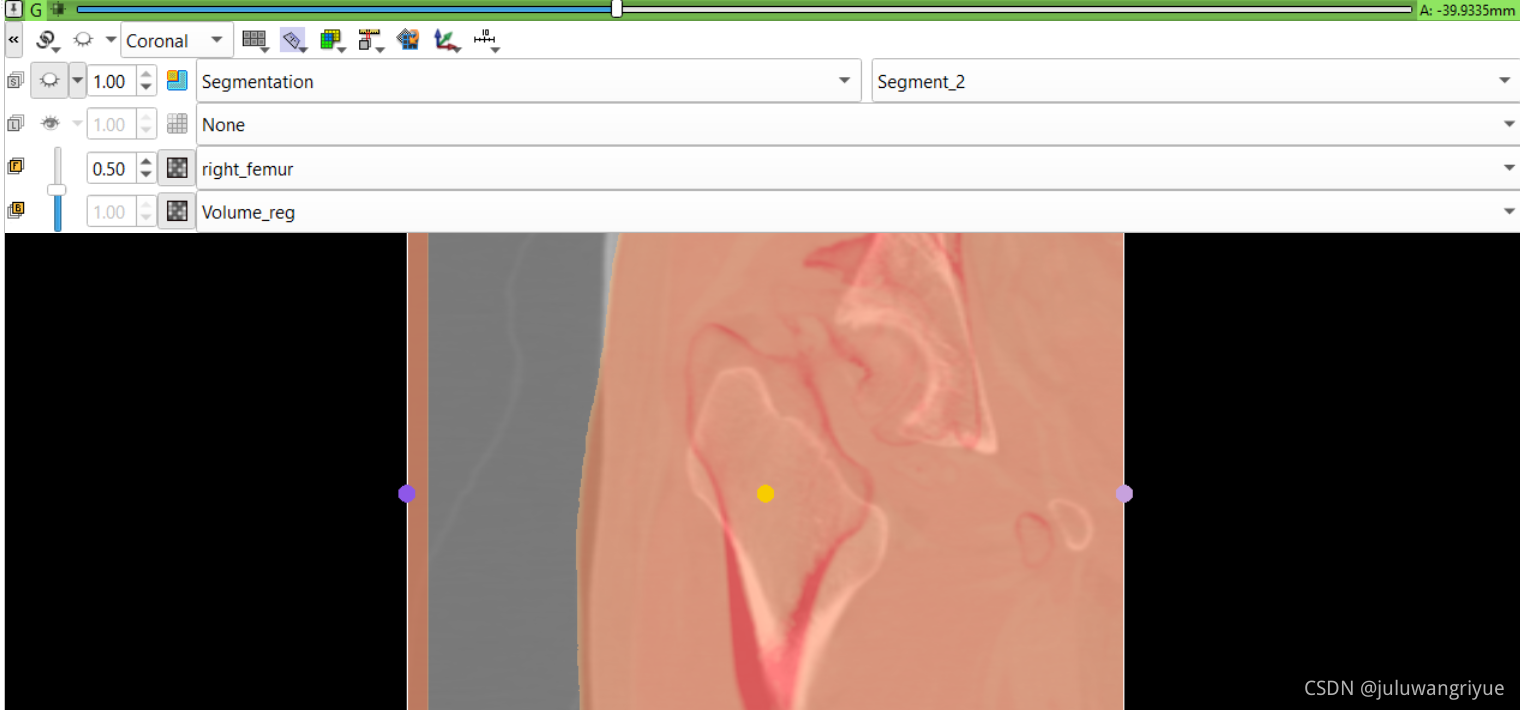
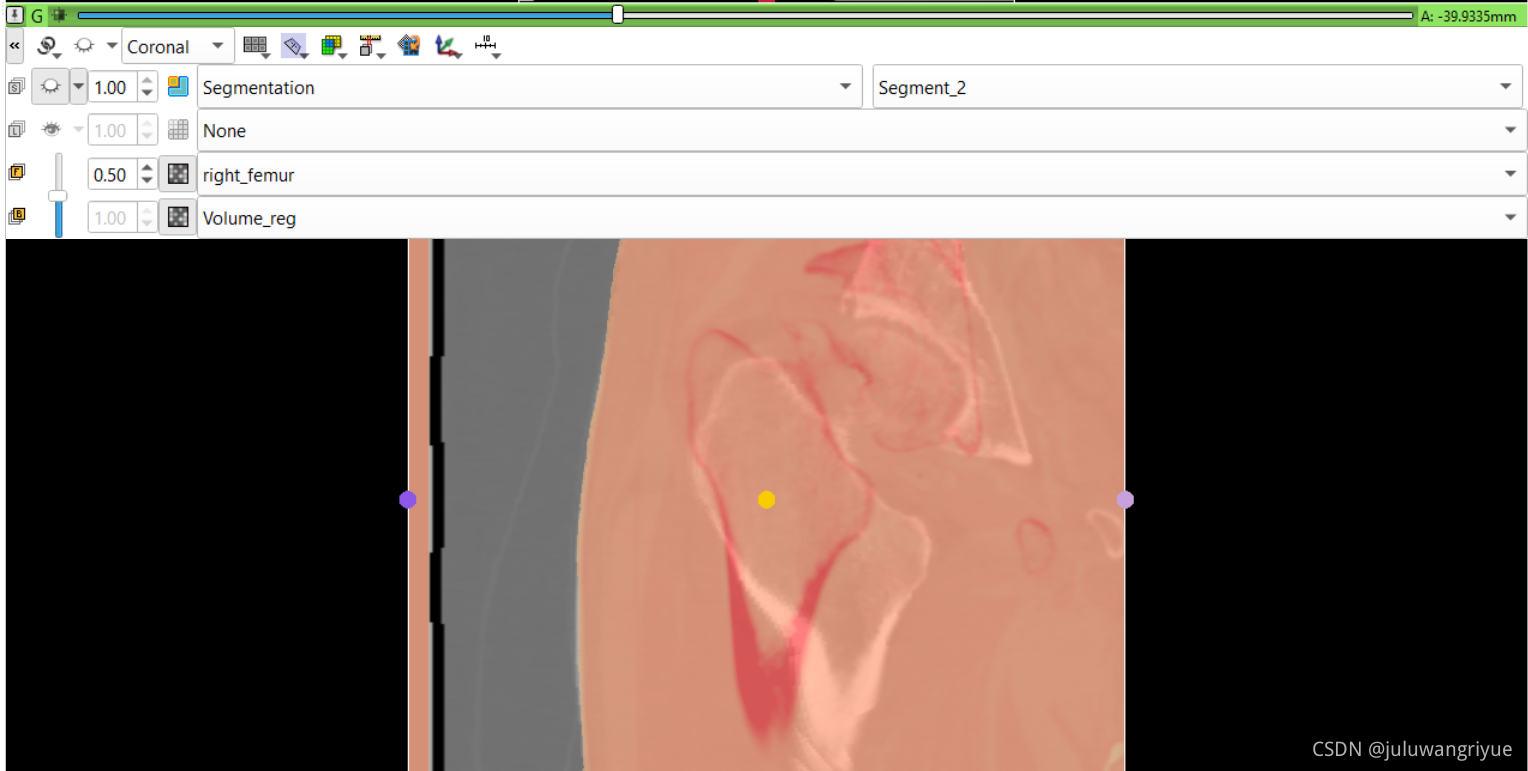
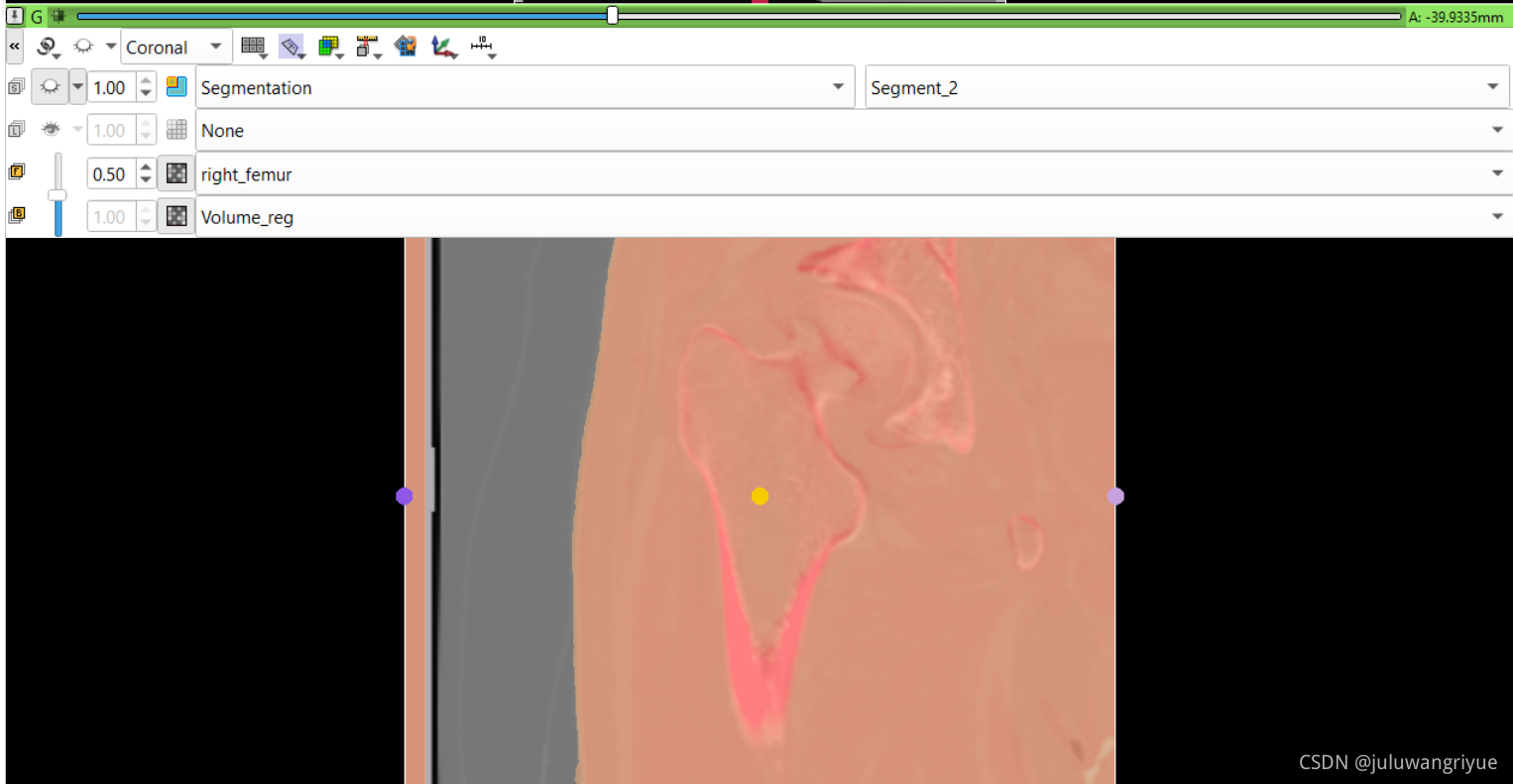
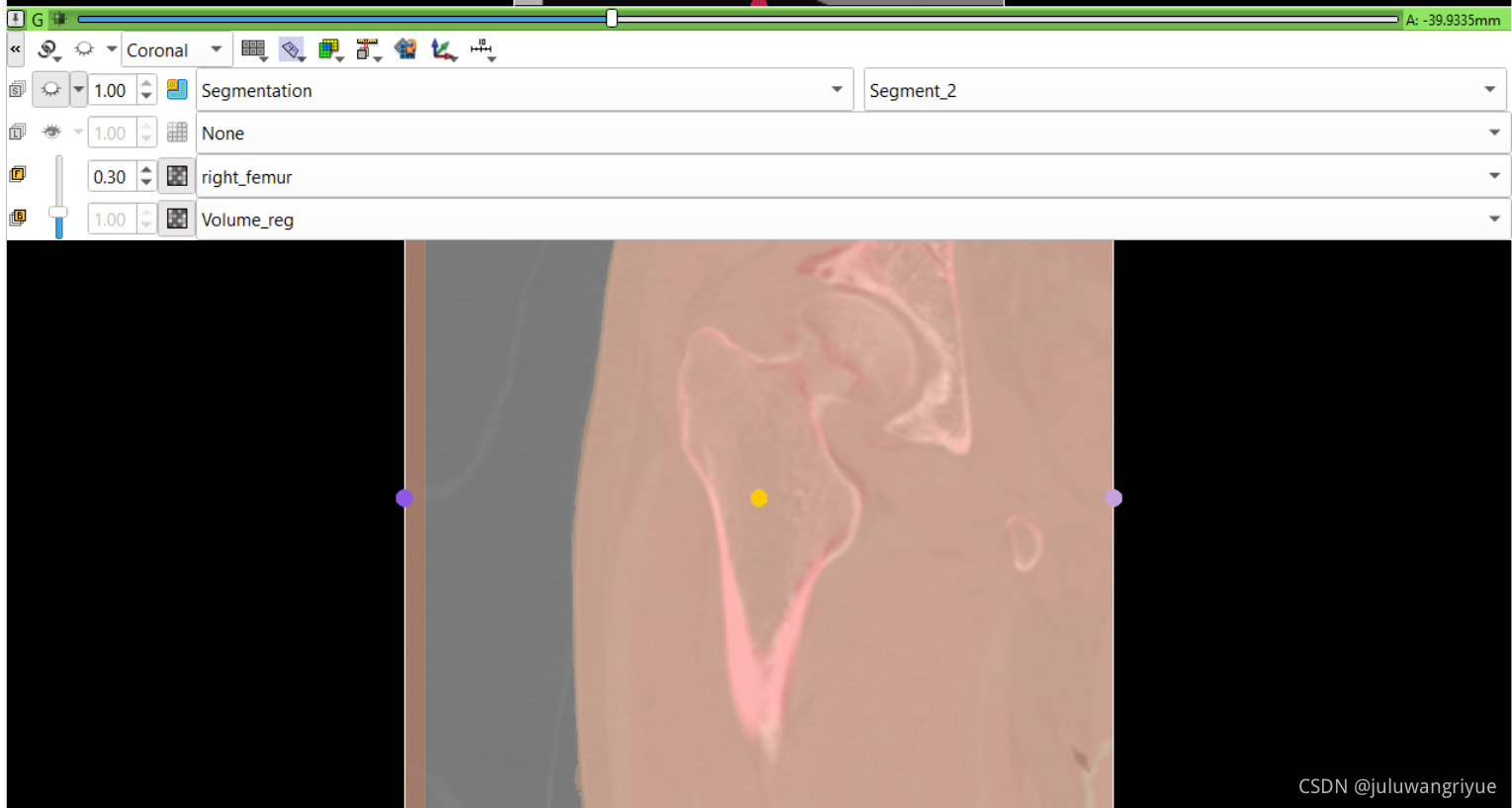
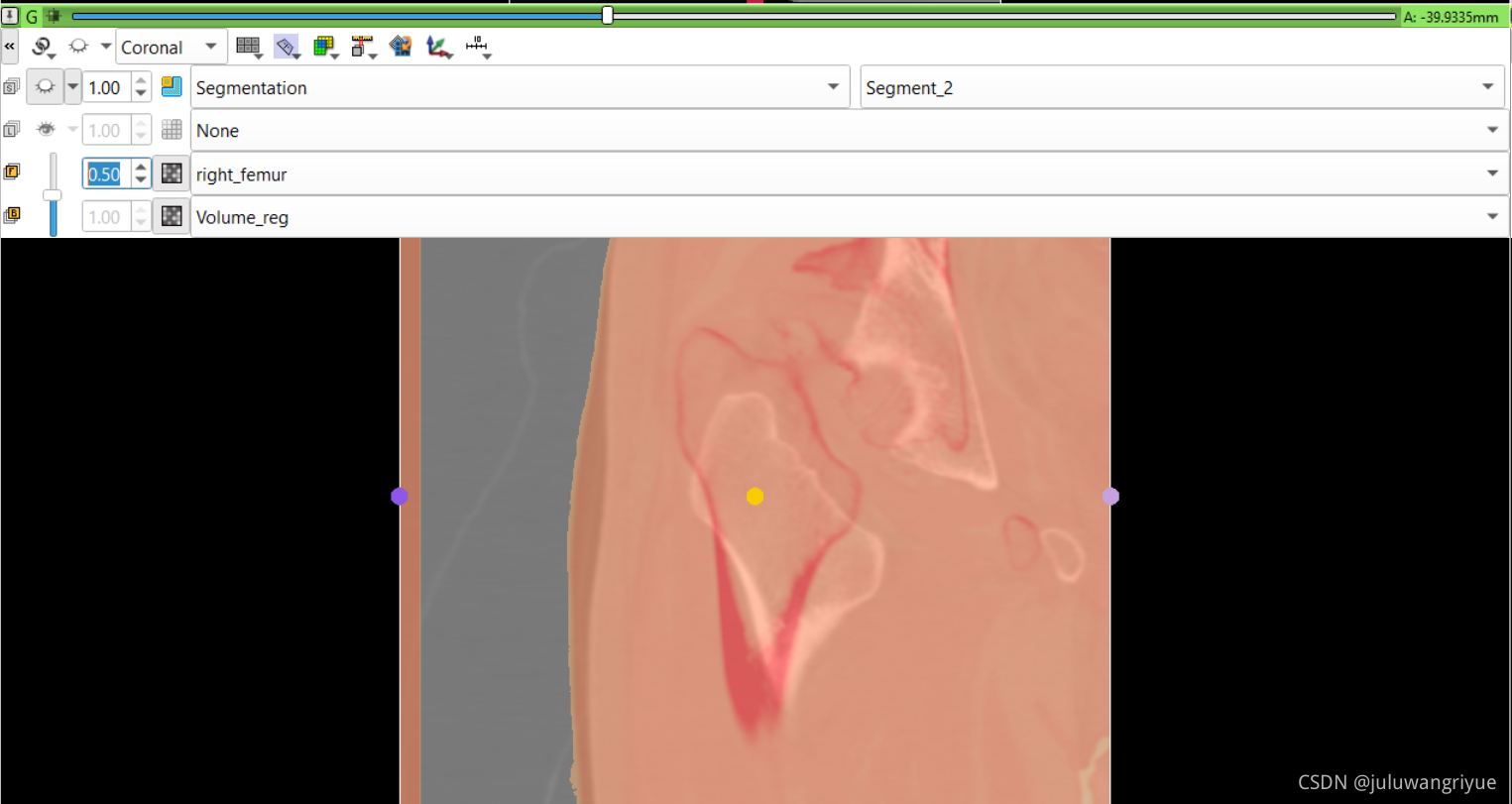
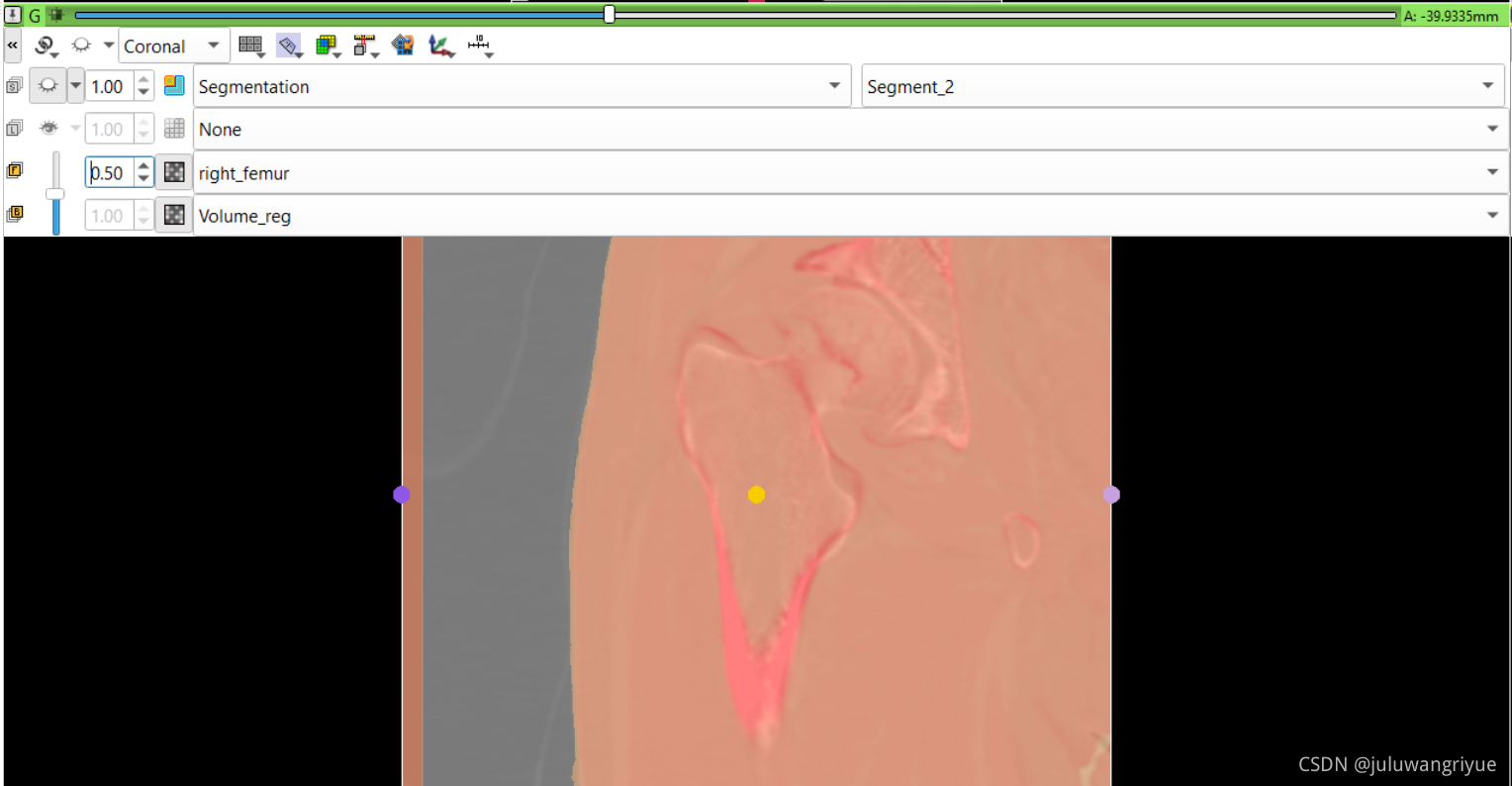
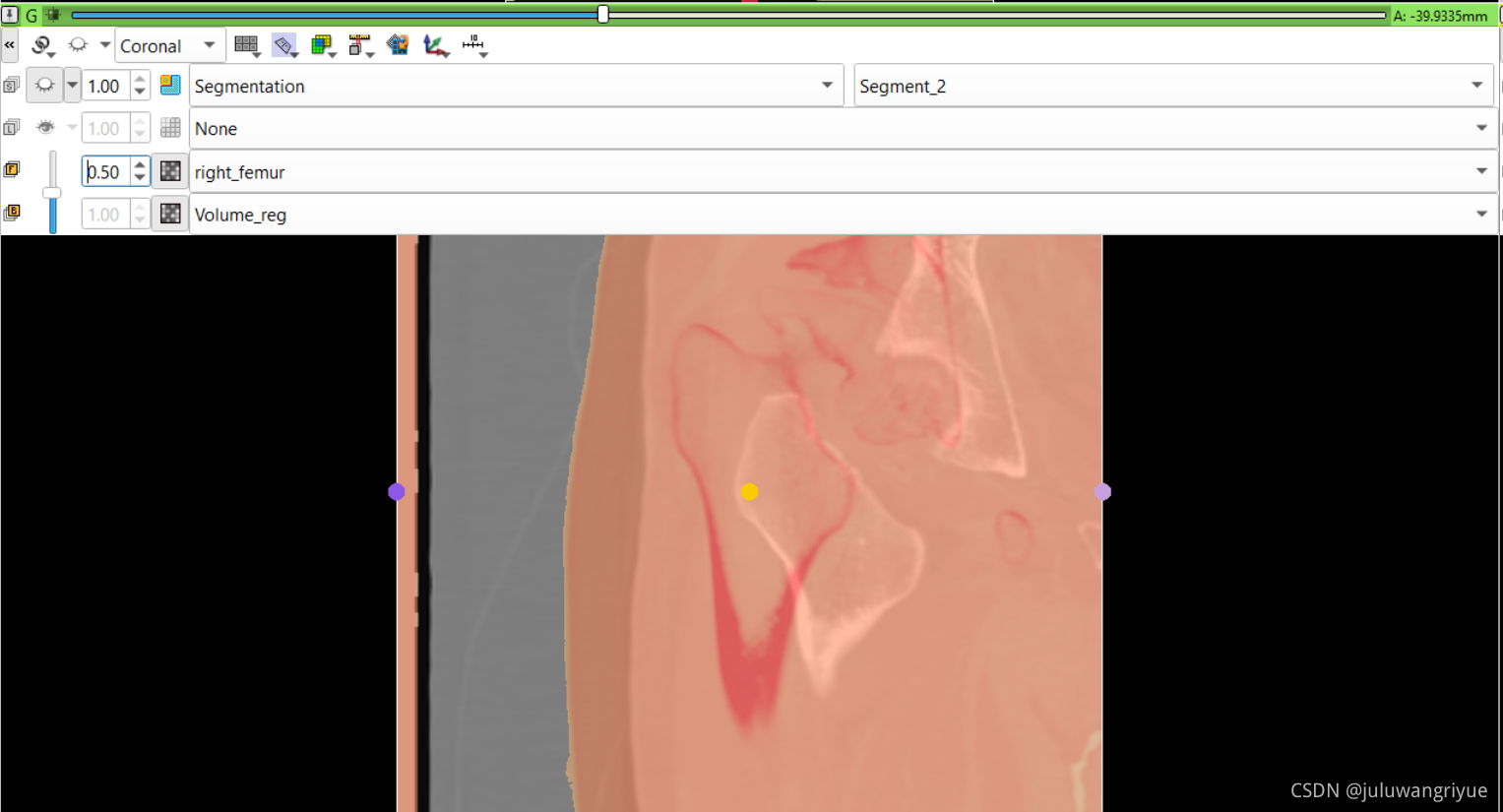
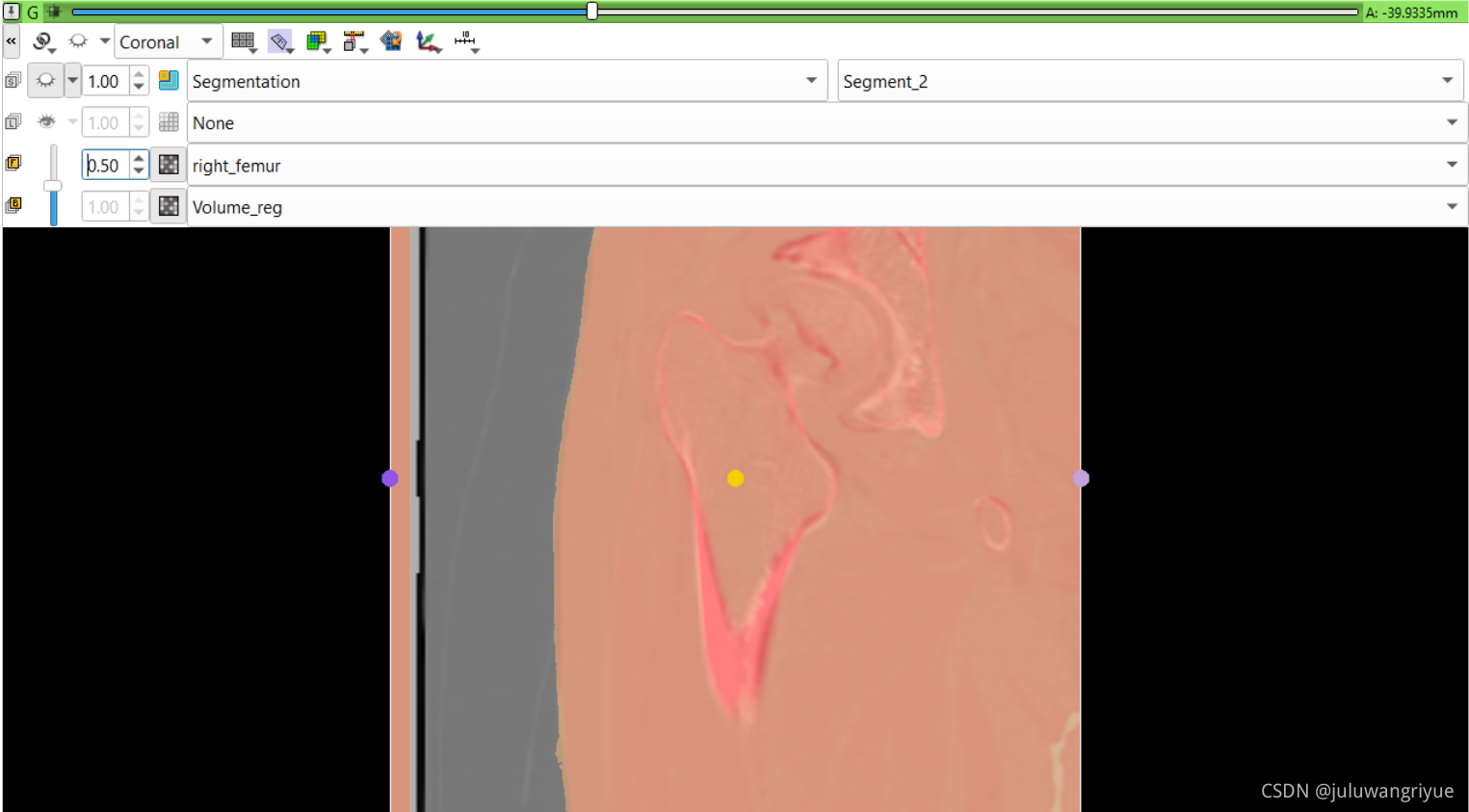
左侧股骨镜像并平移对齐后再配准的图像
配置不同的颜色显示,确认配准效果
配准变换参数
2、尝试不同的配准方法,找最好配准效果的方法
可用方法 31种(重名的3组,5个3D CT, monomodal (lung),2个extremity MRI (wrist),2个3D MR, multi-contrast (rat brain)):
generic (all)
generic rigid (all)
3D MR T1, monomodal (brain)
3D MR BFFE, monomodal (prostate)
3D MRT1 & PET (brain)
3D CT, monomodal (lung)
3D CT, monomodal (lung)
3D MRI, monomodal (brain)
3D MR HASTE, monomodal (brain)
3D CT, monomodal (lung)
3D MRI, monomodal (brain)
3D MR HASTE, monomodal (brain)
3D CT, monomodal (lung)
2D x-ray- 3D CT (cerebral)
3D microCT (whole-body mouse)
3D CT, monomodal (lung)
3D MR, monomodal (knee cartilage)
3D MR, multi sequence (carotid artery)
3D CT, monomodal (head and neck)
3D MR, multi-contrast (rat brain)
3D CT, 3D MR, multimodal (head and neck)
3D MR, multi-contrast (mouse brain)
3D MR, multi-contrast (rat brain)
3D T1 fat-suppressed Ga-enhanced extremity MR (wrist)
3D DCE-MRI (breast)
3D MR (mouse brain)
2D CBF MR (mouse brain)
extremity MRI (wrist)
extremity MRI (wrist)
MRI (spine)
CT/ MR-based pseudo-CT (pelvis (prostate))
3D CT, multi-contrast (cardiac)
效果对比:
generic rigid (all)
效果较差,时间较快不到3分钟
3D MR T1, monomodal (brain)
时间较长,大概十几分钟,CPU几乎占满,内存占用6G~7G,效果比较好
3D MR BFFE, monomodal (prostate)
CPU和内存占用不是很多3G~4G,时间占用大概2分钟,效果较差
3D MRT1 & PET (brain)
内存2G,时间1分钟,效果较差
3D CT, monomodal (lung)
程序崩溃
3D MRI, monomodal (brain)
时间1分钟,内存3G,配准效果不好
3D MR HASTE, monomodal (brain)
内存3.7G,时间2分钟,效果较好
2D x-ray- 3D CT (cerebral)
程序崩溃
3D microCT (whole-body mouse)
内存6G,时间6分钟,结果较好
3D MR, monomodal (knee cartilage)
内存2G,时间小于1分钟,结果不好
3D MR, multi sequence (carotid artery)
程序崩溃
3D CT, monomodal (head and neck)
内存6.4G,CPU占满,时间10分钟,结果较好
3D MR, multi-contrast (rat brain)
内存6.3G,CPU占满,时间20分钟以上,效果不好
3D CT, 3D MR, multimodal (head and neck)
内存3.8G,CPU占满,时间15分钟,效果较好
3D MR, multi-contrast (mouse brain)
程序崩溃
3D T1 fat-suppressed Ga-enhanced extremity MR (wrist)
内存4.5G,CPU占满, 时间10分钟,程序崩溃。
3D DCE-MRI (breast)
生成的是二值图像,对此问题不能使用
3D MR (mouse brain)
内存占满,CPU占满,时间较长,大于50分钟。
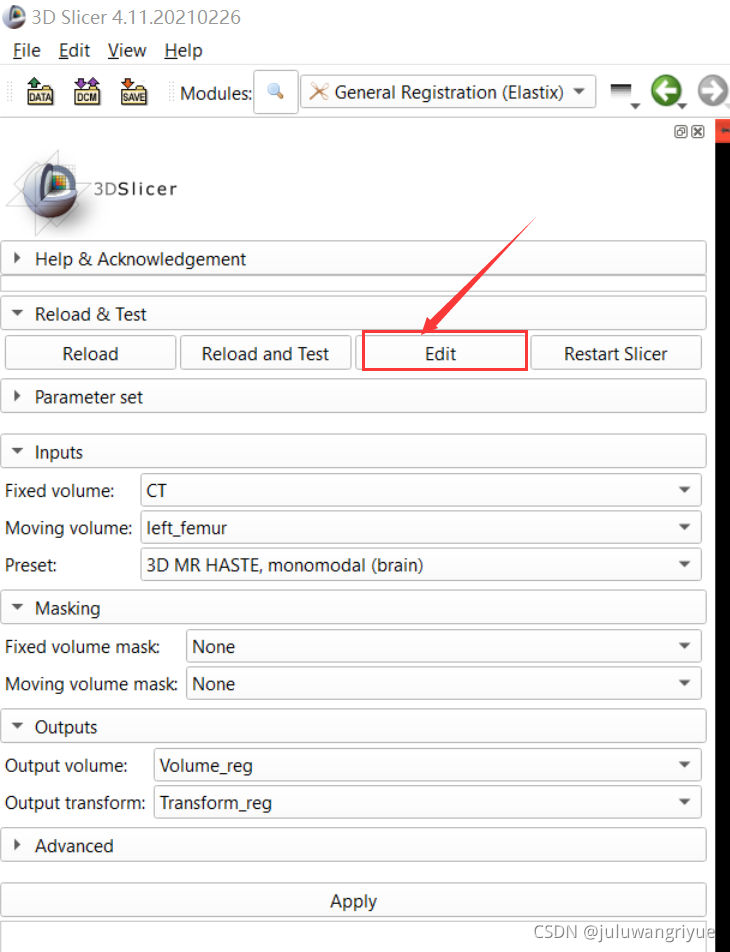
查看各个transform实现,可以通过界面Edit进入python脚本查看实现。主要看metric 评价标准和transform方式。
# 获取矩阵变换信息
3、slicer中Elastix配准的实现
从Edit进入 C:\Users\wmz\AppData\Local\NA-MIC\Slicer 4.11.20210226\NA-MIC\Extensions-29738\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py
可以查看方法的实现,执行配准 只是调用了elastix.exe和transformix.exe,使用获取的参数执行,然后获取结果。而且会生成一些临时变量保存到C:\Users\wmz\AppData\Local\Temp\Slicer\Elastix文件夹下,再读取返回变换后的数据。
获取预设参数
比如选择的预设参数 3D MR HASTE, monomodal (brain) ,会从 C:\Users\wmz\AppData\Local\NA-MIC\Slicer 4.11.20210226\NA-MIC\Extensions-29738\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Resources\RegistrationParameters\ElastixParameterSetDatabase.xml 文件中读取对应的参数
<ParameterSet id="par0010"
modality="3D MR HASTE, monomodal"
content="brain"
description="interpatient; affine + B-spline transformation; mutual information"
publications="Klein (2010) - Early diagnosis of dementia based on intersubject whole-brain dissimilarities">
<ParameterFiles>
<File Name="Par0010affine.txt" />
<File Name="Par0010bspline.txt" />
</ParameterFiles>
选择模式 3D MR HASTE, monomodal (brain) 会用到2个参数文件Par0010affine.txt 和 Par0010bspline.txt。
Par0010affine.txt 文件中的内容包含:图像类型(像素类型:浮点型,图像维度:3维),组件(配准、图像金字塔、插值方式、度量标准、优化器、重采样插值器、重采样器、变换类型等),每一个分辨率水平最大迭代次数(1000次)等。
// Description: affine, MI, ASGD
//ImageTypes
(FixedInternalImagePixelType "float")
(FixedImageDimension 3)
(MovingInternalImagePixelType "float")
(MovingImageDimension 3)
//Components
(Registration "MultiResolutionRegistration")
(FixedImagePyramid "FixedSmoothingImagePyramid")
(MovingImagePyramid "MovingSmoothingImagePyramid")
(Interpolator "BSplineInterpolator")
(Metric "AdvancedMattesMutualInformation")
(Optimizer "AdaptiveStochasticGradientDescent")
(ResampleInterpolator "FinalBSplineInterpolator")
(Resampler "DefaultResampler")
(Transform "AffineTransform")
(ErodeMask "false" )
(NumberOfResolutions 4)
(HowToCombineTransforms "Compose")
(AutomaticTransformInitialization "true")
(AutomaticScalesEstimation "true")
(AutomaticTransformInitializationMethod "CenterOfGravity" )
(WriteTransformParametersEachIteration "false")
(WriteResultImage "false")
(CompressResultImage "true")
(WriteResultImageAfterEachResolution "false")
(ShowExactMetricValue "false")
//Maximum number of iterations in each resolution level:
(MaximumNumberOfIterations 1000)
//Number of grey level bins in each resolution level:
(NumberOfHistogramBins 32 )
(FixedLimitRangeRatio 0.0)
(MovingLimitRangeRatio 0.0)
(FixedKernelBSplineOrder 3)
(MovingKernelBSplineOrder 3)
//Number of spatial samples used to compute the mutual information in each resolution level:
(ImageSampler "Random")
(NumberOfSpatialSamples 2000 )
(NewSamplesEveryIteration "true")
(CheckNumberOfSamples "true")
(MaximumNumberOfSamplingAttempts 10)
//Order of B-Spline interpolation used in each resolution level:
(BSplineInterpolationOrder 1)
//Order of B-Spline interpolation used for applying the final deformation:
(FinalBSplineInterpolationOrder 3)
//Default pixel value for pixels that come from outside the picture:
(DefaultPixelValue 0)
(MaximumStepLength 4.0)
Par0010bspline.txt 文件内容:图像类型(像素类型:浮点型,图像维度:3维),组件(配准、图像金字塔、插值方式、度量标准、优化器、重采样插值器、重采样器、变换类型等),每一个分辨率水平最大迭代次数(2000次)等.
// Description: bspline, localised MI
//ImageTypes
(FixedInternalImagePixelType "float")
(FixedImageDimension 3)
(MovingInternalImagePixelType "float")
(MovingImageDimension 3)
//Components
(Registration "MultiResolutionRegistration")
(FixedImagePyramid "FixedSmoothingImagePyramid")
(MovingImagePyramid "MovingSmoothingImagePyramid")
(Interpolator "BSplineInterpolator")
(Metric "AdvancedMattesMutualInformation")
(Optimizer "StandardGradientDescent")
(ResampleInterpolator "FinalBSplineInterpolator")
(Resampler "DefaultResampler")
(Transform "BSplineTransform")
(ErodeMask "false" )
(NumberOfResolutions 4)
(FinalGridSpacingInPhysicalUnits 15.0 15.0 15.0)
(GridSpacingSchedule 4 4 2 1)
(HowToCombineTransforms "Compose")
(WriteTransformParametersEachIteration "false")
(WriteTransformParametersEachResolution "false")
(WriteResultImage "false")
(CompressResultImage "true")
(WriteResultImageAfterEachResolution "false")
(ShowExactMetricValue "false")
//Maximum number of iterations in each resolution level:
(MaximumNumberOfIterations 2000 )
//Number of grey level bins in each resolution level:
(NumberOfHistogramBins 32 )
(FixedLimitRangeRatio 0.0)
(MovingLimitRangeRatio 0.0)
(FixedKernelBSplineOrder 3)
(MovingKernelBSplineOrder 3)
(UseFastAndLowMemoryVersion "true")
//Number of spatial samples used to compute the mutual information in each resolution level:
(ImageSampler "RandomCoordinate")
(FixedImageBSplineInterpolationOrder 1 )
(UseRandomSampleRegion "true")
(SampleRegionSize 50.0 50.0 50.0)
(NumberOfSpatialSamples 2000 )
(NewSamplesEveryIteration "true")
(CheckNumberOfSamples "true")
(MaximumNumberOfSamplingAttempts 10)
//Order of B-Spline interpolation used in each resolution level:
(BSplineInterpolationOrder 1)
//Order of B-Spline interpolation used for applying the final deformation:
(FinalBSplineInterpolationOrder 3)
//Default pixel value for pixels that come from outside the picture:
(DefaultPixelValue 0)
//SP: Param_a in each resolution level. a_k = a/(A+k+1)^alpha
(SP_a 10000.0 )
//SP: Param_A in each resolution level. a_k = a/(A+k+1)^alpha
(SP_A 100.0 )
//SP: Param_alpha in each resolution level. a_k = a/(A+k+1)^alpha
(SP_alpha 0.6 )
def getRegistrationPresets(self):
if self.registrationPresets:
return self.registrationPresets
# Read database from XML file
elastixParameterSetDatabasePath = os.path.join(self.scriptPath, 'Resources', 'RegistrationParameters', 'ElastixParameterSetDatabase.xml')
if not os.path.isfile(elastixParameterSetDatabasePath):
raise ValueError("Failed to open parameter set database: "+elastixParameterSetDatabasePath)
elastixParameterSetDatabaseXml = vtk.vtkXMLUtilities.ReadElementFromFile(elastixParameterSetDatabasePath)
elastixParameterSetDatabaseXml.UnRegister(None)
# Create python list from XML for convenience
self.registrationPresets = []
for parameterSetIndex in range(elastixParameterSetDatabaseXml.GetNumberOfNestedElements()):
parameterSetXml = elastixParameterSetDatabaseXml.GetNestedElement(parameterSetIndex)
parameterFilesXml = parameterSetXml.FindNestedElementWithName('ParameterFiles')
parameterFiles = []
for parameterFileIndex in range(parameterFilesXml.GetNumberOfNestedElements()):
parameterFiles.append(parameterFilesXml.GetNestedElement(parameterFileIndex).GetAttribute('Name'))
self.registrationPresets.append([parameterSetXml.GetAttribute('id'), parameterSetXml.GetAttribute('modality'),
parameterSetXml.GetAttribute('content'), parameterSetXml.GetAttribute('description'), parameterSetXml.GetAttribute('publications'), parameterFiles])
return self.registrationPresets
执行配准
def registerVolumes(self, fixedVolumeNode, movingVolumeNode, parameterFilenames = None, outputVolumeNode = None, outputTransformNode = None,
fixedVolumeMaskNode = None, movingVolumeMaskNode = None, forceDisplacementFieldOutputTransform = False):
self.abortRequested = False
tempDir = self.createTempDirectory()
self.addLog('Volume registration is started in working directory: '+tempDir)
# Write inputs
inputDir = os.path.join(tempDir, 'input')
qt.QDir().mkpath(inputDir)
inputParamsElastix = []
# Add input volumes
inputVolumes = []
inputVolumes.append([fixedVolumeNode, 'fixed.mha', '-f'])
inputVolumes.append([movingVolumeNode, 'moving.mha', '-m'])
inputVolumes.append([fixedVolumeMaskNode, 'fixedMask.mha', '-fMask'])
inputVolumes.append([movingVolumeMaskNode, 'movingMask.mha', '-mMask'])
for [volumeNode, filename, paramName] in inputVolumes:
if not volumeNode:
continue
# Save original file paths
originalFilePath = ""
originalFilePaths = []
volumeStorageNode = volumeNode.GetStorageNode()
if volumeStorageNode:
originalFilePath = volumeStorageNode.GetFileName()
for fileIndex in range(volumeStorageNode.GetNumberOfFileNames()):
originalFilePaths.append(volumeStorageNode.GetNthFileName(fileIndex))
# Save to new location
filePath = os.path.join(inputDir, filename)
slicer.util.saveNode(volumeNode, filePath, {
"useCompression": False})
inputParamsElastix.append(paramName)
inputParamsElastix.append(filePath)
# Restore original file paths
if volumeStorageNode:
volumeStorageNode.ResetFileNameList()
volumeStorageNode.SetFileName(originalFilePath)
for fileIndex in range(volumeStorageNode.GetNumberOfFileNames()):
volumeStorageNode.AddFileName(originalFilePaths[fileIndex])
else:
# temporary storage node was created, remove it to restore original state
volumeStorageNode = volumeNode.GetStorageNode()
slicer.mrmlScene.RemoveNode(volumeStorageNode)
# Specify output location
resultTransformDir = os.path.join(tempDir, 'result-transform')
qt.QDir().mkpath(resultTransformDir)
inputParamsElastix += ['-out', resultTransformDir]
# Specify parameter files
if parameterFilenames == None:
parameterFilenames = self.getRegistrationPresets()[0][RegistrationPresets_ParameterFilenames]
for parameterFilename in parameterFilenames:
inputParamsElastix.append('-p')
parameterFilePath = os.path.abspath(os.path.join(self.registrationParameterFilesDir, parameterFilename))
inputParamsElastix.append(parameterFilePath)
# Run registration
ep = self.startElastix(inputParamsElastix)
self.logProcessOutput(ep)
# Write results
if not self.abortRequested:
transformFileName = resultTransformDir+'/TransformParameters.'+str(len(parameterFilenames)-1)+'.txt'
#Load Linear Transform if available
elastixTransformFileImported = False
if outputTransformNode and (not forceDisplacementFieldOutputTransform):
try:
transformFromParent = vtk.vtkGeneralTransform()
self.readElastixTransformToVTK(transformFileName, transformFromParent)
# Save transform into transform node (as a simple matrix, if possible)
transformFromParentLinear = vtk.vtkTransform()
if slicer.vtkMRMLTransformNode.IsGeneralTransformLinear(transformFromParent, transformFromParentLinear):
outputTransformNode.SetMatrixTransformFromParent(transformFromParentLinear.GetMatrix())
else:
outputTransformNode.SetAndObserveTransformFromParent(transformFromParent)
elastixTransformFileImported = True
except:
# Could not load transform (probably not linear and bspline)
elastixTransformFileImported = False
#Create temp results directory
resultResampleDir = os.path.join(tempDir, 'result-resample')
qt.QDir().mkpath(resultResampleDir)
inputParamsTransformix = []
inputParamsTransformix += ['-tp', transformFileName]
inputParamsTransformix += ['-out', resultResampleDir]
if outputVolumeNode:
inputParamsTransformix += ['-in', os.path.join(inputDir, 'moving.mha')]
if outputTransformNode:
inputParamsTransformix += ['-def', 'all']
#Run Transformix to get resampled moving volume or transformation as a displacement field
if (outputVolumeNode is not None) or (not elastixTransformFileImported):
tp = self.startTransformix(inputParamsTransformix)
self.logProcessOutput(tp)
if outputVolumeNode:
#Load volume in Slicer
outputVolumePath = os.path.join(resultResampleDir, "result.mhd")
[success, loadedOutputVolumeNode] = slicer.util.loadVolume(outputVolumePath, returnNode = True)
if success:
outputVolumeNode.SetAndObserveImageData(loadedOutputVolumeNode.GetImageData())
ijkToRas = vtk.vtkMatrix4x4()
loadedOutputVolumeNode.GetIJKToRASMatrix(ijkToRas)
outputVolumeNode.SetIJKToRASMatrix(ijkToRas)
slicer.mrmlScene.RemoveNode(loadedOutputVolumeNode)
else:
self.addLog("Failed to load output volume from "+outputVolumePath)
if (outputTransformNode) and (not elastixTransformFileImported):
#Load transform in Slicer
outputTransformPath = os.path.join(resultResampleDir, "deformationField.mhd")
[success, loadedOutputTransformNode] = slicer.util.loadTransform(outputTransformPath, returnNode = True)
if success:
if loadedOutputTransformNode.GetReadAsTransformToParent():
outputTransformNode.SetAndObserveTransformToParent(loadedOutputTransformNode.GetTransformToParent())
else:
outputTransformNode.SetAndObserveTransformFromParent(loadedOutputTransformNode.GetTransformFromParent())
slicer.mrmlScene.RemoveNode(loadedOutputTransformNode)
else:
self.addLog("Failed to load output transform from "+outputTransformPath)
if slicer.app.majorVersion >= 5 or (slicer.app.majorVersion >= 4 and slicer.app.minorVersion >= 11):
outputTransformNode.AddNodeReferenceID(slicer.vtkMRMLTransformNode.GetMovingNodeReferenceRole(), movingVolumeNode.GetID())
outputTransformNode.AddNodeReferenceID(slicer.vtkMRMLTransformNode.GetFixedNodeReferenceRole(), fixedVolumeNode.GetID())
# Clean up
if self.deleteTemporaryFiles:
import shutil
shutil.rmtree(tempDir)
self.addLog("Registration is completed")